| Version: | 4.3.1 |
| Date: | 2025-09-18 |
| Title: | Transparent Assessment Framework for Reproducible Research |
| Imports: | grDevices, lattice, methods, stats, tools, utils |
| LazyData: | yes |
| Description: | General framework to organize data, methods, and results used in reproducible scientific analyses. A TAF analysis consists of four scripts (data.R, model.R, output.R, report.R) that are run sequentially. Each script starts by reading files from a previous step and ends with writing out files for the next step. Convenience functions are provided to version control the required data and software, run analyses, clean residues from previous runs, manage files, manipulate tables, and produce figures. With a focus on stability and reproducible analyses, the TAF package comes with no dependencies. TAF forms a base layer for the 'icesTAF' package and other scientific applications. |
| License: | GPL-3 |
| URL: | https://github.com/ices-tools-prod/TAF |
| Encoding: | UTF-8 |
| RoxygenNote: | 7.3.3 |
| NeedsCompilation: | no |
| Packaged: | 2025-09-18 10:21:02 UTC; arnim |
| Author: | Arni Magnusson [aut, cre], Colin Millar [aut], Iago Mosqueira [aut], Alexandros Kokkalis [ctb], Ibrahim Umar [ctb], Hjalte Parner [ctb] |
| Maintainer: | Arni Magnusson <thisisarni@gmail.com> |
| Repository: | CRAN |
| Date/Publication: | 2025-09-18 12:50:21 UTC |
Transparent Assessment Framework for Reproducible Research
Description
General framework to organize data, methods, and results used in reproducible
scientific analyses. A TAF analysis consists of four scripts (data.R,
model.R, output.R, report.R) that are run sequentially.
Each script starts by reading files from a previous step and ends with
writing out files for the next step.
Convenience functions are provided to version control the required data and software, run analyses, clean residues from previous runs, manage files, manipulate tables, and produce figures. With a focus on stability and reproducible analyses, the TAF package comes with no dependencies. TAF forms a base layer for the icesTAF package and other scientific applications.
The following diagram describes the general workflow of every TAF analysis:
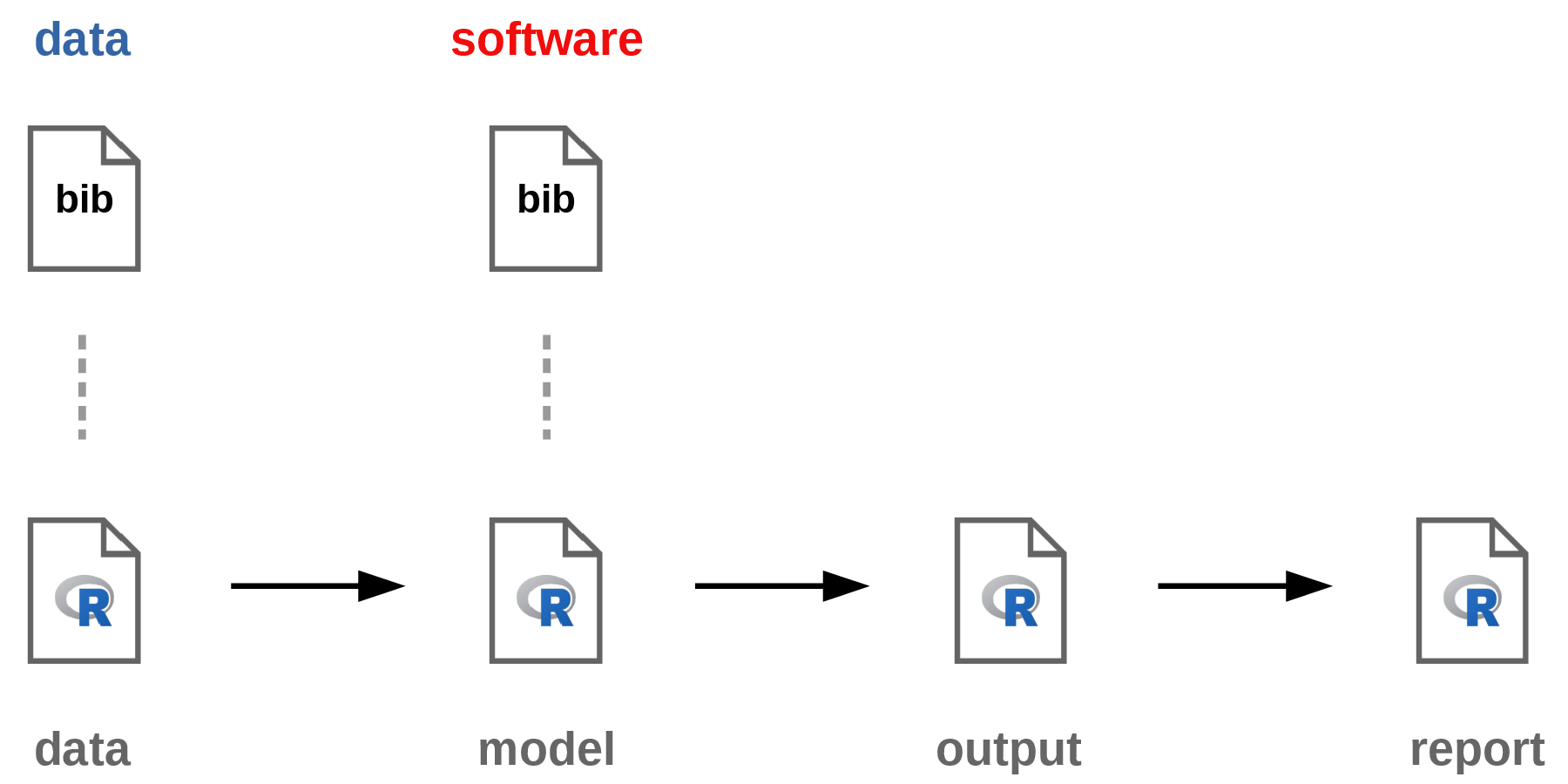
See vignette("TAF") for more detailed coverage on how to organize and
run TAF workflows.
Details
Initial TAF steps:
draft.data | draft DATA.bib file |
draft.software | draft SOFTWARE.bib file |
period | paste period string for DATA.bib |
taf.boot | set up data files and software |
taf.example | copy example analysis from TAF package |
taf.skeleton | create empty TAF template |
Running scripts:
clean | clean TAF directories |
clean.boot | clean boot directory |
make | run R script if needed |
make.all | run all TAF scripts as needed |
make.taf | run TAF script if needed |
msg | show message |
source.all | run all TAF scripts |
source.taf | run TAF script |
File management:
convert.spaces | convert spaces |
cp | copy files |
mkdir | create directory |
os.linux | operating system |
os.macos | operating system |
os.windows | operating system |
read.taf | read CSV file |
source.dir | source all *.R files |
taf.library | load package from TAF library |
taf.unzip | unzip file |
write.taf | write CSV file |
Tables:
ddim | show data dimensions |
div | divide column values |
flr2taf | convert FLR to TAF |
long2taf | convert long format to TAF |
long2xtab | convert long format to crosstab |
plus | rename plus group column |
rnd | round column values |
sam2taf | convert SAM to TAF |
taf2html | convert data frame to HTML |
taf2long | convert TAF to long format |
taf2xtab | convert TAF to crosstab |
tt | transpose TAF table |
wide2long | convert wide to long format |
xtab2long | convert crosstab to long format |
xtab2taf | convert crosstab to TAF |
Plots:
lim | compute axis limits |
taf.colors | predefined colors |
taf.png | open PNG graphics device |
zoom | change lattice text size |
Example tables:
catage.long | long format |
catage.taf | TAF format |
catage.wide | wide format |
catage.xtab | crosstab format |
summary.taf | summary results |
Example workflow:
linreg | simple linear regression |
Administrative or auxiliary tools:
check.software | check global package versions |
clean.data | clean boot data |
clean.library | clean TAF library |
clean.software | clean TAF software |
deps | workflow dependencies |
detach.packages | detach all packages |
dir.tree | show directory structure |
dos2unix | convert line endings |
download | download file |
download.github | download repository |
draft.readme | draft introductory readme |
file.encoding | examine file encoding |
get.remote.sha | look up SHA code |
git.repo | read Git repository name |
install.deps | install dependencies |
is.r.package | check if file is an R package |
latin1.to.utf8 | convert file encoding |
line.endings | examine line endings |
pdeps | package dependencies |
read.bib | read metadata entries |
rmdir | remove empty directory |
taf.boot.path | construct path to boot folder |
taf.data.path | construct path to boot data files |
taf.install | install package in TAF library |
taf.libPaths | add TAF library to search path |
taf.libraries | load all packages from TAF library |
taf.session | show session information |
taf.sources | list metadata entries |
unix2dos | convert line endings |
utf8.to.latin1 | convert file encoding |
Author(s)
Arni Magnusson, Colin Millar, and Iago Mosqueira.
See Also
See vignette("TAF") for an introduction to the TAF package.
The TAF Wiki provides additional help resources.
Examples
## Not run:
taf.example("linreg")
setwd("linreg")
taf.boot()
source.all()
## End(Not run)
Various TAF Helper Functions
Description
These functions are called by user-level functions. The functionality is documented in the user-level functions.
Usage
boot.dir()
boot.dir.inside(inside = ".")
boot.exists()
extract.subdir(targz, subtargz, subdir)
stamp.description(targz, spec, sha.full)
ds.file(package, author, year, title, version, source)
ds.package(package, author, year, title, version, source)
model.dir()
model.script()
parse.repo(repo)
process.bibfile(type, clean = TRUE, quiet = FALSE, ...)
process.entry(bib, clean = FALSE, comma.split = TRUE, force = FALSE,
quiet = FALSE)
already.in.taf.library(targz, lib)
taf.pkg()
See Also
TAF-package gives an overview of the package.
Catch at Age in Long Format
Description
Small catch-at-age table to describe a long format data frame to store year-age values.
Usage
catage.long
Format
Data frame containing three columns:
Year | year |
Age | age |
Catch | catch (millions of individuals) |
Details
The data are an excerpt (first years and ages) from the catch-at-age table for North Sea cod from the ICES (2016) assessment.
Source
ICES (2016). Report of the working group on the assessment of demersal stocks in the North Sea and Skagerrak (WGNSSK). ICES CM 2016/ACOM:14, p. 673. doi:10.17895/ices.pub.5329.
See Also
catage.taf and catage.xtab describe alternative
table formats.
long2taf converts a long table to TAF format.
TAF-package gives an overview of the package.
Examples
catage.long
long2taf(catage.long)
Catch at Age in TAF Format
Description
Small catch-at-age table to describe a TAF format data frame to store year-age values.
Usage
catage.taf
Format
Data frame containing five columns:
Year | year |
1 | number of one-year-olds in the catch (millions) |
2 | number of two-year-olds in the catch (millions) |
3 | number of three-year-olds in the catch (millions) |
4 | number of four-year-olds in the catch (millions) |
Details
The data are an excerpt (first years and ages) from the catch-at-age table for North Sea cod from the ICES (2016) assessment.
Source
ICES (2016). Report of the working group on the assessment of demersal stocks in the North Sea and Skagerrak (WGNSSK). ICES CM 2016/ACOM:14, p. 673. doi:10.17895/ices.pub.5329.
See Also
catage.long and catage.xtab describe alternative
table formats.
taf2long and taf2xtab convert a TAF table to
alternative formats.
wide2long converts a wide table to long format.
TAF-package gives an overview of the package.
Examples
catage.taf
taf2long(catage.taf)
taf2xtab(catage.taf)
Catch at Age in Wide Format
Description
Catch-at-age table to describe a wide format data frame to store area-year-age values.
Usage
catage.wide
Format
Data frame containing six columns:
Area | area |
Year | year |
1 | number of one-year-olds in the catch (millions) |
2 | number of two-year-olds in the catch (millions) |
3 | number of three-year-olds in the catch (millions) |
4 | number of four-year-olds in the catch (millions) |
Details
The data are an excerpt (first years and ages) from the catch-at-age table for North Sea cod from the ICES (2016) assessment. Catches in ‘area 1’ are the original data, while ‘area 2’ contains the same values multiplied by two.
Source
ICES (2016). Report of the working group on the assessment of demersal stocks in the North Sea and Skagerrak (WGNSSK). ICES CM 2016/ACOM:14, p. 673. doi:10.17895/ices.pub.5329.
See Also
catage.taf describes the TAF format.
taf2long converts a TAF table to long format.
TAF-package gives an overview of the package.
Examples
catage.wide
wide2long(catage.wide)
Catch at Age in Crosstab Format
Description
Small catch-at-age table to describe a crosstab format data frame to store year-age values.
Usage
catage.xtab
Format
Data frame with years as row names and containing four columns:
1 | number of one-year-olds in the catch (millions) |
2 | number of two-year-olds in the catch (millions) |
3 | number of three-year-olds in the catch (millions) |
4 | number of four-year-olds in the catch (millions) |
Details
The data are an excerpt (first years and ages) from the catch-at-age table for North Sea cod from the ICES (2016) assessment.
Source
ICES (2016). Report of the working group on the assessment of demersal stocks in the North Sea and Skagerrak (WGNSSK). ICES CM 2016/ACOM:14, p. 673. doi:10.17895/ices.pub.5329.
See Also
catage.long and catage.taf describe alternative
table formats.
xtab2taf converts a crosstab table to TAF format.
TAF-package gives an overview of the package.
Examples
catage.xtab
xtab2taf(catage.xtab)
Check SOFTWARE.bib Against Global Packages
Description
Compare versions declared in SOFTWARE.bib with packages installed in
the global R library.
Usage
check.software(full = FALSE)
Arguments
full |
whether to return full data frame as output. |
Value
Logical vector (or data frame if full = TRUE) indicating which
installed packages are ready, i.e., at least as new as the version
required in SOFTWARE.bib.
A warning is generated if any installed packages are older than required.
Note
Generally, TAF installs R packages that are declared in SOFTWARE.bib
inside the TAF library (boot/library). This guarantees that the right
versions of packages are installed for the analysis. The taf.library
function is then used to load packages from the TAF library.
In special cases, however, it might be useful to compare the versions of
packages declared in SOFTWARE.bib against packages that are installed
in the global R library, outside the TAF library.
See Also
taf.boot and taf.library are the general tools to
install and load packages of the correct version in the TAF library.
update.packages can be used to update packages in the general R
library to the newest version available on CRAN.
Examples
## Not run:
check.software()
check.software(full=TRUE)
## End(Not run)
Clean TAF Directories
Description
Remove TAF directories: data, model, output, and
report.
Usage
clean(dirs = c("data", model.dir(), "output", "report"), force = FALSE)
Arguments
dirs |
directories to delete. |
force |
passed to |
Details
The model directory may also be named method and is cleaned in
the same way.
Value
No return value, called for side effects.
Note
The purpose of removing the directories is to make sure that subsequent TAF scripts start by creating new empty directories.
If any of the dirs is "boot", it is treated specially and
clean.boot is called to clean the boot directory.
In other words, clean("boot") and clean.boot() are
interchangeable, the latter providing a slightly clearer interface that was
introduced in version 4.2.0.
See Also
clean.boot cleans the boot directory.
mkdir and rmdir create and remove empty
directories.
TAF-package gives an overview of the package.
Examples
## Not run:
clean()
clean.boot()
## End(Not run)
Clean Boot Directory
Description
Clean the boot directory using clean.data, clean.library, and
clean.software.
Usage
clean.boot(force = FALSE)
Arguments
force |
passed to |
Value
No return value, called for side effects.
Note
Instead of completely removing the boot directory, clean.data,
clean.library, and clean.software are used to clean the
boot/data, boot/library, and boot/library
subdirectories. This protects the subdirectory boot/initial, boot
scripts, and *.bib metadata files from being accidentally deleted.
See Also
clean cleans TAF directories: data, model,
output, and report.
clean.data selectively removes data from boot/data.
clean.library selectively removes packages from
boot/library.
clean.software selectively removes software from
boot/software.
TAF-package gives an overview of the package.
Examples
## Not run:
clean()
clean.boot()
## End(Not run)
Clean Data
Description
Selectively remove data from the boot/data folder if not listed in
DATA.bib.
Usage
clean.data(folder = "boot/data", quiet = FALSE, force = FALSE)
Arguments
folder |
location of |
quiet |
whether to suppress messages about removed data. |
force |
whether to remove |
Value
No return value, called for side effects.
Note
For each data file or subfolder, the cleaning procedure selects between two cases:
Data entry found in
DATA.bib- do nothing.Data entry is not listed in
DATA.bib- remove.
The taf.boot procedure cleans the boot/data folder, without
requiring the user to run clean.data.
See Also
taf.boot calls clean.data as part of the default boot
procedure.
clean.software cleans the local TAF software folder.
clean.library cleans the local TAF library.
TAF-package gives an overview of the package.
Examples
## Not run:
clean.data()
## End(Not run)
Clean TAF Library
Description
Selectively remove packages from the local TAF library if not listed in
SOFTWARE.bib.
Usage
clean.library(folder = "boot/library", quiet = FALSE, force = FALSE)
Arguments
folder |
location of local TAF library. |
quiet |
whether to suppress messages about removed packages. |
force |
whether to remove the local TAF library, regardless of how it
compares to |
Value
No return value, called for side effects.
Note
For each package, the cleaning procedure selects between three cases:
Installed package matches
SOFTWARE.bib- do nothing.Installed package is not the version listed in
SOFTWARE.bib- remove.Installed package is not listed in
SOFTWARE.bib- remove.
The taf.boot procedure cleans the TAF library, without requiring the
user to run clean.library. The main reason for a TAF user to run
clean.library directly is to experiment with installing and removing
different versions of software without modifying the SOFTWARE.bib
file.
See Also
taf.boot calls clean.library as part of the default boot
procedure.
taf.install installs a package in the local TAF library.
clean.software cleans the local TAF software folder.
clean.data cleans the boot/data folder.
TAF-package gives an overview of the package.
Examples
## Not run:
clean.library()
## End(Not run)
Clean TAF Software
Description
Selectively remove software from the local TAF software folder if not listed
in SOFTWARE.bib.
Usage
clean.software(folder = "boot/software", quiet = FALSE, force = FALSE)
Arguments
folder |
location of local TAF software folder. |
quiet |
whether to suppress messages about removed software. |
force |
whether to remove the local TAF software folder, regardless of
how it compares to |
Value
No return value, called for side effects.
Note
For each file (and subdirectory) in the software folder, the cleaning procedure selects between three cases:
File and version matches
SOFTWARE.bib- do nothing.Filename does not contain the version listed in
SOFTWARE.bib- remove.File is not listed in
SOFTWARE.bib- remove.
The taf.boot procedure cleans the TAF software folder, without
requiring the user to run clean.software. The main reason for a TAF
user to run clean.software directly is to experiment with installing
and removing different versions of software without modifying the
SOFTWARE.bib file.
See Also
taf.boot calls clean.software as part of the default
boot procedure.
download.github downloads a GitHub repository.
clean.library cleans the local TAF library.
clean.data cleans the boot/data folder.
TAF-package gives an overview of the package.
Examples
## Not run:
clean.software()
## End(Not run)
Convert Spaces
Description
Convert spaces in filenames.
Usage
convert.spaces(file, sep = "_")
Arguments
file |
filename, e.g. |
sep |
character to use instead of spaces. |
Value
TRUE for success, FALSE for failure, invisibly.
Note
This function treats ‘%20’ in filenames as a space and converts to
sep.
See Also
file.rename is the base function to rename files.
TAF-package gives an overview of the package.
Examples
## Not run:
write(pi, "A B.txt")
convert.spaces("A B.txt")
# Many files
convert.spaces("boot/initial/data/*")
## End(Not run)
Copy Files
Description
Copy or move files, overwriting existing files if necessary, and returning the result invisibly.
Usage
cp(from, to, move = FALSE, ignore = FALSE, overwrite = TRUE,
quiet = TRUE)
Arguments
from |
source filenames, e.g. |
to |
destination filenames, or directory. |
move |
whether to move instead of copy. |
ignore |
whether to suppress error if source file does not exist. |
overwrite |
whether to overwrite if destination file exists. |
quiet |
whether to suppress messages. |
Value
TRUE for success, FALSE for failure, invisibly.
Note
To prevent accidental loss of files, two safeguards are enforced when
move = TRUE:
When moving files, the
toargument must either have a filename extension or be an existing directory.When moving many files to one destination, the
toargument must be an existing directory.
If these conditions do not hold, no files are changed and an error is returned.
See Also
file.copy and unlink are the underlying functions
used to copy and (if move = TRUE) delete files.
file.rename is the base function to rename files.
TAF-package gives an overview of the package.
Examples
## Not run:
write(pi, "A.txt")
cp("A.txt", "B.txt")
cp("A.txt", "B.txt", move=TRUE)
# Copy directory tree
cp(system.file(package="datasets"), ".")
mkdir("everything")
cp("datasets/*", "everything")
## End(Not run)
Data Dimensions
Description
Show the data dimensions of a table.
Usage
ddim(x, reduce = FALSE)
Arguments
x |
a data frame where the first columns are dimension variables and the last column is a measurement variable. |
reduce |
is whether to omit single-level dimensions. |
Details
Dimension variables can include year, age, region, fleet, survey, or the like, generally an integer or string. The measurement variable can be catch, fishing mortality, maturity, weight, or the like, often a decimal.
x can also be an FLR table in FLQuant format.
Value
Named vector showing the dimension names and number of levels.
See Also
unique is the base function to extract the levels of a
dimension variable.
TAF-package gives an overview of the package.
Examples
# Long table format, 8 years and 4 ages
ddim(catage.long)
# Some base R datasets
ddim(esoph[-5])
ddim(rev(warpbreaks))
ddim(rev(ToothGrowth))
Dependencies of a Workflow
Description
Search R scripts for packages that are required.
Usage
deps(path = ".", base = FALSE, installed = TRUE, available = TRUE,
list = FALSE)
Arguments
path |
a directory or file containing R code. |
base |
whether to include base packages in the output. |
installed |
whether to include installed packages in the output. |
available |
whether to include available packages in the output. |
list |
whether to return packages in list format, split by script. |
Details
The files analyzed are those with the file extensions .R, .r,
.Rmd, and .rmd.
Value
Names of packages as a vector, or in list format if list=TRUE. If no
dependencies are found, the return value is NULL.
Note
Package names are matched based on four patterns:
library(*) require(*) *::object *:::object
The search algorithm may return false-positive dependencies if these patterns occur inside if-clauses, strings, comments, etc.
See Also
installed.packages, available.packages.
pdeps shows the dependencies of a package.
TAF-package gives an overview of the package.
Examples
## Not run:
dir <- system.file(package="MASS", "scripts")
script <- system.file(package="MASS", "scripts/ch08.R")
deps(script) # dependencies
deps(script, base=TRUE) # including base packages
deps(script, installed=FALSE) # not (yet) installed
deps(dir)
deps(dir, list=TRUE)
deps(dir, available=FALSE) # dependencies that might be unavailable
## End(Not run)
Detach Packages
Description
Detach all non-base packages that have been attached using library or
taf.library.
Usage
detach.packages(quiet = FALSE)
Arguments
quiet |
whether to suppress messages. |
Value
Names of detached packages.
See Also
detach is the underlying base function to detach a package.
taf.library loads a package from boot/library.
TAF-package gives an overview of the package.
Examples
## Not run:
detach.packages()
## End(Not run)
Directory Tree
Description
Show directory structure and files in a tree format.
Usage
dir.tree(path = ".")
Arguments
path |
the directory to show. |
See Also
dir is the underlying base function that returns directories and files as strings.
TAF-package gives an overview of the package.
Examples
## Not run:
path <- system.file("examples", package="TAF")
dir.tree(path)
cbind(dir(recursive=TRUE, include.dirs=TRUE))
## End(Not run)
Divide Columns
Description
Divide column values in a data frame with a common number.
Usage
div(x, cols, by = 1000, grep = FALSE, ...)
Arguments
x |
a data frame. |
cols |
column names, or column indices. |
by |
a number to divide with. |
grep |
whether |
... |
passed to |
Value
A data frame similar to x, after dividing columns cols by the
number by.
Note
Provides notation that is convenient for modifying many columns at once.
See Also
transform can also be used to recalculate column values, using
a more general and verbose syntax.
grep is the underlying function used to match column names if
grep is TRUE.
rnd is a similar function that rounds columns.
TAF-package gives an overview of the package.
Examples
# These are equivalent:
x1 <- div(summary.taf, c("Rec","Rec_lo","Rec_hi",
"TSB","TSB_lo","TSB_hi",
"SSB","SSB_lo","SSB_hi",
"Removals","Removals_lo","Removals_hi"))
x2 <- div(summary.taf, "Rec|TSB|SSB|Removals", grep=TRUE)
x3 <- div(summary.taf, "Year|Fbar", grep=TRUE, invert=TRUE)
# Less reliable in scripts if columns have been added/deleted/reordered:
x4 <- div(summary.taf, 2:13)
Convert Line Endings
Description
Convert line endings in a text file between Dos (CRLF) and Unix (LF) format.
Usage
dos2unix(file, force = FALSE)
unix2dos(file, force = FALSE)
Arguments
file |
a filename. |
force |
whether to proceed with the conversion when the file is not a standard text file. |
Details
The default value of force = FALSE is a safety feature that can avoid
corrupting files that are not standard text files, such as binary files. A
standard text file is one that can be read using readLines
without producing warnings.
Value
No return value, called for side effects.
See Also
line.endings examines line endings.
TAF-package gives an overview of the package.
Examples
## Not run:
file <- "test.txt"
write("123", file)
dos2unix(file)
file.size(file)
unix2dos(file)
file.size(file)
file.remove(file)
## End(Not run)
Download File
Description
Download a file in binary mode, e.g. a model executable.
Usage
download(url, dir = ".", mode = "wb", chmod = file_ext(url) == "",
destfile = file.path(dir, basename(url)), quiet = TRUE, ...)
Arguments
url |
URL of file to download. |
dir |
directory to download to. |
mode |
download mode, see details. |
chmod |
whether to set execute permission (default is |
destfile |
destination path and filename (optional, overrides
|
quiet |
whether to suppress messages. |
... |
passed to |
Details
With the default mode "wb" the file is downloaded in binary mode (see
download.file), to prevent R from adding ^M at line
ends. This is particularly relevant for Windows model executables, while the
chmod switch is useful when downloading Linux executables.
This function can be convenient for downloading any file, including text
files. Data files in CSV or other text format can also be read directly into
memory using read.table, read.taf or similar functions, without
writing to the file system.
Value
No return value, called for side effects.
Note
If destfile contains a question mark it is removed from the
destfile filename. Similarly, if destfile contains spaces or
‘%20’ sequences, those are converted to underscores.
In general, TAF scripts do not access the internet using
download or similar functions. Instead, data and software are declared
in DATA.bib and SOFTWARE.bib and then downloaded using
taf.boot. The exception is when a boot script is used to fetch
files from a web service (see
TAF Wiki).
See Also
download.file is the underlying base function to download
files.
download.github downloads a GitHub repository.
TAF-package gives an overview of the package.
Examples
## Not run:
url <- paste0("https://github.com/ices-taf/2015_had-iceg/raw/master/",
"bootstrap/initial/software/catageysa/catageysa.exe")
download(url)
## End(Not run)
Download GitHub Repository
Description
Download a repository from GitHub in ‘tar.gz’ format.
Usage
download.github(repo, dir = ".", quiet = FALSE)
Arguments
repo |
GitHub reference of the form |
dir |
directory to download to. |
quiet |
whether to suppress messages. |
Value
Name of downloaded tar.gz file.
Note
In general, TAF scripts do not access the internet using
download.github or similar functions. Instead, data and software are
declared in DATA.bib and SOFTWARE.bib and then downloaded using
taf.boot. The exception is when a boot script is used to fetch
files from a web service (see
TAF Wiki).
See Also
taf.boot uses download.github to fetch software and data
repositories.
download downloads a file.
untar extracts a tar.gz archive.
taf.install installs a package in tar.gz format.
TAF-package gives an overview of the package.
Examples
## Not run:
# Specify release tag
download.github("ices-tools-prod/icesAdvice@1.3-0")
# Specify SHA reference code
download.github("ices-tools-prod/icesAdvice@4271797")
## End(Not run)
Draft DATA.bib
Description
Create an initial draft version of a ‘DATA.bib’ metadata file.
Usage
draft.data(originator = NULL, year = format(Sys.time(), "%Y"),
title = NULL, period = NULL, access = "Public", source = NULL,
file = "", append = FALSE,
data.files = dir(taf.boot.path("initial/data")),
data.scripts = dir(boot.dir(), pattern = "\\.R$"))
Arguments
originator |
who prepared the data, e.g. a working group acronym. |
year |
year of the analysis when the data were used. The default is the current year. |
title |
description of the data, including survey names or the like. |
period |
a string of the form |
access |
data access code: |
source |
where the data are copied/downloaded from. This can be a URL,
filename, or a special value: |
file |
optional filename to save the draft metadata to a file. The value
|
append |
whether to append metadata entries to an existing file. |
data.files |
data files to consider. The default is all folders and
files inside |
data.scripts |
boot data scripts to consider. The default is all
|
Details
Typical usage is to specify originator, while using the default values
for the other arguments. Most data files have the same originator, which can
be specified to facilitate completing the entries after creating the initial
draft.
The data access codes come from https://vocab.ices.dk/?ref=1435.
The special values source = "file", source = "folder", and
source = "script" are described on the
TAF Wiki, along with
other metadata information.
The default value file = "" prints the initial draft in the console,
instead of writing it to a file. The output can then be pasted into a file to
edit further, without accidentally overwriting an existing metadata file.
Value
Character vector of class Bibtex.
Note
This function is intended to be called from the top directory of a TAF
analysis. It looks for data files inside boot/initial/data folder and
data scripts inside boot.
After creating the initial draft, the user can complete the description of
each data entry inside the title field and look into each file to
specify the period that the data cover.
See Also
period pastes two years to form a period string.
draft.software creates an initial draft version of a
SOFTWARE.bib metadata file.
taf.boot reads and processes metadata entries.
TAF-package gives an overview of the package.
Examples
## Not run:
# Print in console
draft.data("WGEF", 2015)
# Export to file
draft.data("WGEF", 2015, file=TRUE)
# Empty entry, to complete by hand
draft.data(data.files="")
## End(Not run)
Draft Readme
Description
Draft an introductory README.md that describes how to run a TAF
analysis.
Usage
draft.readme(title = NULL, file = "")
Arguments
title |
title to display at the top of the 'README.md' page. |
file |
optional filename to save the draft readme to a file. The value
|
Details
The default value title = NULL uses the Git repository name as a
placeholder title.
The default value file = "" prints the initial draft in the console,
instead of writing it to a file. The output can then be pasted into a file to
edit further, without accidentally overwriting an existing file.
Value
Character vector of class Bibtex.
Note
Although the output is Markdown text, the Bibtex class is used for
convenient display in the console.
See Also
git.repo reads the Git repository name.
TAF-package gives an overview of the package.
Examples
## Not run:
# Print in console
draft.readme()
# Export to file
draft.readme(file=TRUE)
# Specify title
draft.readme("Yellowfin tuna in the western and central Pacific")
## End(Not run)
Draft SOFTWARE.bib
Description
Create an initial draft version of a ‘SOFTWARE.bib’ metadata file.
Usage
draft.software(package, author = NULL, year = NULL, title = NULL,
version = NULL, source = NULL, file = "", append = FALSE)
Arguments
package |
name of one or more R packages, or files/folders starting with
the path |
author |
author(s) of the software. |
year |
year when this version of the software was released, or the publication year of the cited manual/article/etc. |
title |
title or short description of the software. |
version |
string to specify details about the version, e.g. GitHub branch and commit date. |
source |
string to specify where the software are copied/downloaded
from. This can be a GitHub reference of the form
|
file |
optional filename to save the draft metadata to a file. The value
|
append |
whether to append metadata entries to an existing file. |
Details
Typical usage is to specify package, while using the default values
for the other arguments.
If package is an R package, it can either be a package that is already
installed ("icesAdvice") or a GitHub reference
("ices-tools-prod/icesAdvice@4271797").
With the default version = NULL, the function will automatically
suggest an appropriate version entry for CRAN packages, but for GitHub
packages it is left to the user to add further information about the GitHub
branch (if different from master) and the commit date.
With the default source = NULL, the function will automatically
suggest an appropriate source entry for CRAN and GitHub packages, but for
other R packages it is left to the user to add information about where the
software can be accessed.
The default value file = "" prints the initial draft in the console,
instead of writing it to a file. The output can then be pasted into a file to
edit further, without accidentally overwriting an existing metadata file.
Value
Character vector of class Bibtex.
Note
After creating the initial draft, the user can complete the version,
source, and other fields as required.
This function is especially useful for citing exact versions of R packages on GitHub. To prepare metadata for software other than R packages, see the TAF Wiki for an example.
See Also
citation and packageDescription are the
underlying functions to access information about installed R packages.
draft.data creates an initial draft version of a
DATA.bib metadata file.
taf.boot reads and processes metadata entries.
TAF-package gives an overview of the package.
Examples
# Print in console
draft.software("TAF")
## Not run:
# Export to file
draft.software("TAF", file=TRUE)
## End(Not run)
File Encoding
Description
Examine file encoding.
Usage
file.encoding(file)
Arguments
file |
a filename. |
Value
"latin1", "UTF-8", "unknown", or NA.
This function requires the file shell command to be in the path.
Otherwise, this function returns NA.
Note
The encoding "unknown" indicates that the file is an ASCII text file
or a binary file.
In TAF, text files that have non-ASCII characters should be encoded as UTF-8.
If this function fails in Windows, the guess_encoding function in the
readr package may help.
See Also
Encoding examines the encoding of a string.
latin1.to.utf8 converts files from latin1 to
UTF-8 encoding.
line.endings examines line endings.
TAF-package gives an overview of the package.
Examples
## Not run:
file.base <- system.file(package="base", "DESCRIPTION")
file.nlme <- system.file(package="nlme", "DESCRIPTION")
file.encoding(file.base) # ASCII
file.encoding(file.nlme)
## End(Not run)
Convert FLR Table to TAF Format
Description
Convert a table from FLR format to TAF format.
Usage
flr2taf(x, colname = "Value")
Arguments
x |
a table of class |
colname |
a column name to use if the FLR table contains only one row. |
Value
A data frame in TAF format.
Note
FLR uses the FLQuant class to store tables as 6-dimensional arrays,
while TAF tables are stored as data frames with a year column.
See Also
catage.taf describes the TAF format.
as.data.frame is a method provided by the FLCore package
to convert FLQuant tables to a 7-column long format.
TAF-package gives an overview of the package.
Examples
x <- array(t(catage.xtab), dim=c(4,8,1,1,1,1))
dimnames(x) <- list(age=1:4, year=1963:1970,
unit="unique", season="all", area="unique", iter=1)
flr2taf(x)
x1 <- x[1,,,,,,drop=FALSE]
flr2taf(x1)
flr2taf(x1, "Juveniles")
Get Remote SHA
Description
Look up SHA reference code on GitHub.
Usage
get.remote.sha(owner, repo, ref, seven = TRUE)
Arguments
owner |
repository owner. |
repo |
repository name. |
ref |
reference. |
seven |
whether to truncate SHA reference code to seven characters. |
Value
SHA reference code as a string.
See Also
taf.boot uses get.remote.sha to determine whether it is
necessary to remove or download files, via clean.library,
clean.software, and download.github.
TAF-package gives an overview of the package.
Examples
## Not run:
get.remote.sha("ices-tools-prod", "icesAdvice", "master")
get.remote.sha("ices-tools-prod", "icesAdvice", "1.3-0")
get.remote.sha("ices-tools-prod", "icesAdvice", "1.3-0", seven=FALSE)
## End(Not run)
Git Repo
Description
Read the Git repository name from the .git folder.
Usage
git.repo(path = ".", owner = FALSE, warn = FALSE)
Arguments
path |
top directory of a Git repository. |
owner |
whether to include the repository owner name. |
warn |
whether to generate a warning if no |
Value
String of the format "[owner]/repo".
See Also
draft.readme calls git.repo to include the repository
name in the README.md.
TAF-package gives an overview of the package.
Examples
## Not run:
git.repo()
git.repo(owner=TRUE)
## End(Not run)
Install Dependencies
Description
Search R scripts for packages that are required and install those that are
not already installed. The default install location is the same as
install.packages.
Usage
install.deps(path = ".", ...)
Arguments
path |
a directory or file containing R code. |
... |
passed to |
Details
This function also looks in the TAF boot directory for packages that are required by the TAF boot process, i.e., called from a boot script.
In addition it runs taf.boot on SOFTWARE.bib to install any special packages that may not be available on CRAN.
See Also
install.packages is the underlying function to install
packages.
deps searches R scripts for packages that are required.
TAF-package gives an overview of the package.
Examples
## Not run:
# Download a TAF analysis
download(file.path("https://github.com/ices-taf/2019_san.sa.6",
"archive/refs/heads/master.zip"))
unzip("master.zip")
setwd("2019_san.sa.6-master")
# List dependencies
deps()
deps(taf.boot.path())
# Install dependencies that are not already installed
install.deps()
## End(Not run)
Is R Package
Description
Check if ‘.tar.gz’ file is an R package.
Usage
is.r.package(targz, spec = NULL, warn = TRUE)
Arguments
targz |
a filename ending with |
spec |
an optional list generated with |
warn |
whether to warn if the file contents look like an R package nested inside a repository. |
Details
The only purpose of passing spec is to get a more helpful warning
message if the file contents look like an R package nested inside a
repository.
Value
Logical indicating whether targz is an R package.
Examples
## Not run:
is.r.package("boot/software/SAM.tar.gz")
is.r.package("boot/software/stockassessment.tar.gz")
## End(Not run)
Convert File Encoding
Description
Convert file encoding between "latin1" and "UTF-8".
Usage
latin1.to.utf8(file, force = FALSE)
utf8.to.latin1(file, force = FALSE)
Arguments
file |
a filename. |
force |
whether to perform the conversion even if the current file
encoding cannot be verified with |
Value
No return value, called for side effects.
Note
In TAF, text files that have non-ASCII characters must be encoded as UTF-8.
See Also
iconv converts the encoding of a string.
file.encoding examines the encoding of a file.
TAF-package gives an overview of the package.
Examples
## Not run:
utf8.to.latin1("data.txt")
latin1.to.utf8("data.txt")
## End(Not run)
Axis Limits
Description
Compute reasonable axis limits for plotting non-negative numbers.
Usage
lim(x, mult = 1.1)
Arguments
x |
a vector of data values. |
mult |
a number to multiply with the highest data value. |
Value
A vector of length two, which can be used as axis limits.
Note
The lower limit is set to 0, and the upper limit is determined by the highest data value, times a multiplier.
See Also
TAF-package gives an overview of the package.
Examples
plot(precip)
plot(precip, ylim=lim(precip))
plot(precip, ylim=lim(precip), yaxs="i")
Line Endings
Description
Examine whether file has Dos or Unix line endings.
Usage
line.endings(file)
Arguments
file |
a filename. |
Value
String indicating the line endings: "Dos" or "Unix".
See Also
file.encoding examines the encoding of a file.
dos2unix and unix2dos convert line endings.
TAF-package gives an overview of the package.
Examples
## Not run:
file <- system.file(package="TAF", "DESCRIPTION")
line.endings(file)
## End(Not run)
Linear Regression
Description
Minimal TAF workflow, a simple linear regression where the x and y coordinates come from a text file.
Before the workflow is run, it consists of four scripts and a boot
folder:
[boot] data.R model.R output.R report.R
After the workflow is run, four new folders contain the output from the TAF scripts:
[boot] [data] [model] [output] [report] data.R model.R output.R report.R
See Also
cars describes the dataset.
taf.example copies an example analysis from the TAF package.
TAF-package gives an overview of the package.
Examples
## Not run:
taf.example("linreg")
setwd("linreg")
taf.boot()
source.all()
## End(Not run)
Convert Long Table to TAF Format
Description
Convert a table from long format to TAF format.
Usage
long2taf(x)
Arguments
x |
a data frame in long format. |
Value
A data frame in TAF format.
Note
TAF stores tables as data frames, usually with a year column as seen in stock assessment reports. The long format is more convenient for analysis and producing plots.
See Also
catage.long and catage.taf describe the long and
TAF formats.
taf2long converts a TAF table to long format.
TAF-package gives an overview of the package.
Examples
long2taf(catage.long)
Convert Long Table to Crosstab Format
Description
Convert a table from long format to crosstab format.
Usage
long2xtab(x)
Arguments
x |
a data frame in long format. |
Value
A data frame with years as row names.
See Also
catage.long and catage.xtab describe the long and
crosstab formats.
long2taf and taf2xtab are the underlying
functions that perform the conversion.
TAF-package gives an overview of the package.
Examples
long2xtab(catage.long)
Run R Script if Needed
Description
Run an R script if underlying files have changed, otherwise do nothing.
Usage
make(recipe, prereq, target, include = TRUE, engine = source,
details = FALSE, force = FALSE, recon = FALSE, quiet = TRUE, ...)
Arguments
recipe |
script filename. |
prereq |
one or more files required by the script. For example, data
files, scripts, or |
target |
one or more output files produced by the script. Directory names can also be used. |
include |
whether to automatically include the script itself as a prerequisite file. This means that if the script file has been modified, it should be run. |
engine |
function to source the script. |
details |
whether to show a diagnostic table of files and time last modified. |
force |
whether to run the R script unconditionally. |
recon |
whether to return |
quiet |
whether to suppress messages. |
... |
passed to |
Details
A make() call has the general form
make("analysis.R", "input.dat", "output.dat")
which can be read aloud as:
“script x uses y to produce z”
Value
TRUE or FALSE, indicating whether the script was run.
Note
This function provides functionality similar to makefile rules, to determine whether a script should be (re)run or not.
If any target is either missing or is older than any prereq,
then the script is run.
References
Stallman, R. M. et al. An introduction to makefiles. Chapter 2 in the GNU Make manual.
See Also
source runs any R script, source.taf is more
convenient for running a TAF script, and source.all runs all
TAF scripts.
make, make.taf, and make.all are
similar to the source functions, except they avoid repeating tasks
that have already been run.
TAF-package gives an overview of the package.
The makeit package provides a similar make function, along with
a vignette containing annotated examples and a discussion.
Examples
## Not run:
# Here, model.R uses input.dat, creating results.dat
make("model.R", "data/input.dat", "model/results.dat")
make("model.R", "data/input.dat", "model/results.dat", quiet=FALSE)
make("model.R", "data/input.dat", "model/results.dat", details=TRUE)
## End(Not run)
Run All TAF Scripts as Needed
Description
Run core TAF scripts that have changed, or if previous steps were rerun.
Usage
make.all(...)
Arguments
... |
passed to |
Value
Logical vector indicating which scripts were run.
Note
TAF scripts that will be run as needed are: utilities.R,
data.R, model.R, output.R, and report.R.
The model.R script may also be named method.R and is treated in
the same way.
See Also
source runs any R script, source.taf is more
convenient for running a TAF script, and source.all runs all
TAF scripts.
make, make.taf, and make.all are
similar to the source functions, except they avoid repeating tasks
that have already been run.
TAF-package gives an overview of the package.
Examples
## Not run:
make.all()
## End(Not run)
Run TAF Script if Needed
Description
Run a TAF script if the target directory is either older than the script, or older than the directory of the previous TAF step.
Usage
make.taf(script, ...)
Arguments
script |
TAF script filename. |
... |
passed to |
Value
TRUE or FALSE, indicating whether the script was run.
Note
Any underlying scripts are automatically included if they share the same
filename prefix, followed by an underscore. For example, when determining
whether a script data.R should be run, this function checks whether
data_foo.R and data_bar.R have been recently modified.
See Also
source runs any R script, source.taf is more
convenient for running a TAF script, and source.all runs all
TAF scripts.
make, make.taf, and make.all are
similar to the source functions, except they avoid repeating tasks
that have already been run.
TAF-package gives an overview of the package.
Examples
## Not run:
make.taf("model.R")
## End(Not run)
Create Directory
Description
Create directory, including parent directories if necessary, without generating a warning if the directory already exists.
Usage
mkdir(path)
Arguments
path |
a directory name. |
Value
TRUE for success, FALSE for failure, invisibly.
See Also
dir.create is the base function to create a new directory.
rmdir removes an empty directory.
clean can be used to remove non-empty directories.
TAF-package gives an overview of the package.
Examples
## Not run:
mkdir("emptydir")
rmdir("emptydir")
mkdir("outer/inner")
rmdir("outer", recursive=TRUE)
## End(Not run)
Show Message
Description
Show a message, as well as the current time.
Usage
msg(...)
Arguments
... |
passed to |
Value
No return value, called for side effects.
See Also
message is the base function to show messages, without the
current time.
source.taf reports progress using msg.
TAF-package gives an overview of the package.
Examples
msg("script.R running...")
Operating System
Description
Determine operating system name.
Usage
os()
os.linux()
os.macos()
os.windows()
os.unix()
Value
os returns the name of the operating system, typically "Linux",
"Darwin", or "Windows".
os.linux, os.macos, os.unix, and os.windows
return TRUE or FALSE.
Note
The macOS operating system identifies itself as "Darwin".
Both Linux and macOS are os.unix.
These shorthand functions can be useful when writing workaround solutions in platform-independent scripts.
See Also
Sys.info is the underlying function used to extract the
operating system name.
TAF-package gives an overview of the package.
Examples
os()
os.linux()
os.macos()
os.unix()
os.windows()
Dependencies of a Package
Description
Find dependencies or reverse dependencies of a CRAN package.
Usage
pdeps(packages, recursive = TRUE, reverse = FALSE, base = FALSE,
installed = TRUE, available = TRUE, sort = FALSE, ...)
Arguments
packages |
package names. |
recursive |
whether to include dependencies of dependencies. |
reverse |
whether to find reverse dependencies instead. |
base |
whether to include base packages. |
installed |
whether to include installed packages. |
available |
whether to include available packages. |
sort |
whether to sort package dependencies. |
... |
passed to |
Value
Names of packages that are required by package.
See Also
package_dependencies is the underlying base function to find
CRAN package dependencies.
installed.packages, available.packages.
deps shows the dependencies of a workflow.
TAF-package gives an overview of the package.
Examples
## Not run:
# TAF dependencies
pdeps("TAF") # does not depend on non-base packages
pdeps("TAF", base=TRUE) # depends on these base packages
pdeps("TAF", reverse=TRUE) # icesTAF depends on TAF
# Other packages with light dependencies
sapply(pdeps(c("data.table", "Rcpp", "renv")), length)
# ggplot2 dependencies
pdeps("ggplot2") # full list of dependencies
pdeps("ggplot2", recursive=FALSE) # primary dependencies
# Each ggplot2 dependency brings in these secondary dependencies
pdeps(pdeps("ggplot2", recursive=FALSE)$ggplot2)
## End(Not run)
Period
Description
Paste two years to form a period string.
Usage
period(x, y = NULL)
Arguments
x |
the first year, vector of years, matrix, or data frame. |
y |
the last year, if |
Details
If x is a vector or a data frame, then the lowest and highest years
are used, and y is ignored.
If x is a matrix or data frame, this function looks for years in the
first column. If the values of the first column do not look like years (four
digits), then it looks for years in the row names.
Value
A string of the form "1990-2000".
Note
This function can be useful when working with draft.data.
See Also
paste is the underlying function to paste strings.
draft.data has an argument called period.
TAF-package gives an overview of the package.
Examples
period(1963, 1970)
period(c(1963, 1970))
period(1963:1970)
period(range(catage.taf$Year))
period(catage.taf$Year)
period(catage.taf)
period(catage.xtab)
period(catage.long)
Rename Plus Group Column
Description
Rename the last column in a data frame, by appending a "+" character.
This is useful if the last column is a plus group.
Usage
plus(x)
Arguments
x |
a data frame. |
Value
A data frame similar to x, after renaming the last column.
Note
If the last column name already ends with a "+", the original data
frame is returned without modifications.
See Also
names is the underlying function to rename columns.
TAF-package gives an overview of the package.
Examples
catage <- catage.taf
# Rename last column
catage <- plus(catage)
# Shorter and less error-prone than
names(catage)[names(catage)=="4"] <- "4+"
Read Metadata Entries
Description
Read metadata entries written in BibTeX format.
Usage
read.bib(file)
Arguments
file |
‘*.bib’ file to parse. |
Value
List of metadata entries.
Note
This function was created when the bibtex package was temporarily removed from CRAN. The current implementation reduces the TAF package dependencies to base R and nothing else.
This parser is similar to the read.bib function in the bibtex
package, except:
It returns a plain list instead of class
bibentry.The fields
bibtypeandkeyare stored as list elements instead of attributes.
See the TAF Wiki page on bib entries.
See Also
taf.boot reads and processes metadata entries.
taf.sources reads metadata entries and adds a type
field.
TAF-package gives an overview of the package.
Examples
## Not run:
bib <- read.bib("DATA.bib")
str(bib)
## End(Not run)
Read TAF Table
Description
Read from a CSV file into a data frame.
Usage
read.taf(file, check.names = FALSE, stringsAsFactors = FALSE,
fileEncoding = "UTF-8", ...)
Arguments
file |
a filename. |
check.names |
whether to enforce regular column names, e.g. convert
column name |
stringsAsFactors |
whether to import strings as factors. |
fileEncoding |
character encoding of input file. |
... |
passed to |
Details
Alternatively, file can be a directory or a vector of filenames, to
read many tables in one call.
Value
A data frame, or a list of data frames if file is a directory or a
vector of filenames.
Note
This function gives a warning when column names are missing or duplicated. It also gives a warning if the data frame has zero rows.
See Also
read.csv is the underlying function used to read a table from a
file.
write.taf writes a data frame to a CSV file.
TAF-package gives an overview of the package.
Examples
## Not run:
write.taf(catage.taf, "catage.csv")
catage <- read.taf("catage.csv")
write.taf(catage)
file.remove("catage.csv")
## End(Not run)
Remove Empty Directory
Description
Remove empty directory under any operating system.
Usage
rmdir(path, recursive = FALSE)
Arguments
path |
a directory name. |
recursive |
whether to remove empty subdirectories as well. |
Value
TRUE for success, FALSE for failure, invisibly.
Note
The base function unlink(dir, recursive=FALSE) does not remove empty
directories in Windows and unlink(dir, recursive=TRUE) removes
non-empty directories, making it unsuitable for tidying up empty ones.
See Also
unlink with recursive = TRUE removes non-empty
directories.
mkdir creates a new directory.
clean can be used to remove non-empty directories.
TAF-package gives an overview of the package.
Examples
## Not run:
mkdir("emptydir")
rmdir("emptydir")
mkdir("outer/inner")
rmdir("outer", recursive=TRUE)
## End(Not run)
Round Columns
Description
Round column values in a data frame.
Usage
rnd(x, cols, digits = 0, grep = FALSE, ...)
Arguments
x |
a data frame. |
cols |
column names, or column indices. |
digits |
number of decimal places. |
grep |
whether |
... |
passed to |
Value
A data frame similar to x, after rounding columns cols to the
number of digits.
Note
Provides notation that is convenient for modifying many columns at once.
See Also
round is the underlying function used to round numbers.
grep is the underlying function used to match column names if
grep is TRUE.
div is a similar function that divides columns with a common
number.
TAF-package gives an overview of the package.
The icesAdvice package provides the icesRound function to round
values for ICES advice sheets.
Examples
# With rnd() we no longer need to repeat the column names:
m <- mtcars
m[c("mpg","disp","qsec")] <- round(m[c("mpg","disp","qsec")])
m <- rnd(m, c("mpg","disp","qsec"))
# The x1/x2/x3/x4 approaches are equivalent:
x1 <- rnd(summary.taf, c("Rec","Rec_lo","Rec_hi",
"TSB","TSB_lo","TSB_hi",
"SSB","SSB_lo","SSB_hi",
"Removals","Removals_lo","Removals_hi"))
x1 <- rnd(x1, c("Fbar","Fbar_lo","Fbar_hi"), 3)
x2 <- rnd(summary.taf, "Rec|TSB|SSB|Removals", grep=TRUE)
x2 <- rnd(x2, "Fbar", 3, grep=TRUE)
x3 <- rnd(summary.taf, "Fbar", grep=TRUE, invert=TRUE)
x3 <- rnd(x3, "Fbar", 3, grep=TRUE)
# Less reliable in scripts if columns have been added/deleted/reordered:
x4 <- rnd(summary.taf, 2:13)
x4 <- rnd(x4, 14:16, 3)
Convert SAM Table to TAF Format
Description
Convert a table from SAM format to TAF format.
Usage
sam2taf(x, colname = NULL, year = TRUE)
Arguments
x |
a matrix containing columns |
colname |
a descriptive column name for the output. |
year |
whether to include a year column. |
Details
The default when colname = NULL is to try to infer a column name from
the x argument. For example,
sam2taf(ssbtable(fit)) sam2taf(ssb) sam2taf(SSB)
will recognize ssbtable calls and ssb object names, implicitly
setting colname = "SSB" if the user does not pass an explicit value
for colname.
Value
A data frame in TAF format.
Note
The stockassessment package provides accessor functions that return a
matrix with columns Estimate, Low, and High, while TAF
tables are stored as data frames with a year column.
See Also
summary.taf describes the TAF format.
catchtable, fbartable, rectable, ssbtable, and
tsbtable (in the stockassessment package) return matrices with
SAM estimates and confidence limits.
The summary method for sam objects produces a summary table
with some key quantities of interest, containing duplicated column names
(Low, High) and rounded values.
TAF-package gives an overview of the package.
Examples
# Example objects
x <- as.matrix(summary.taf[grep("SSB", names(summary.taf))])
rec <- as.matrix(summary.taf[grep("Rec", names(summary.taf))])
tsb <- as.matrix(summary.taf[grep("TSB", names(summary.taf))])
dimnames(x) <- list(summary.taf$Year, c("Estimate", "Low", "High"))
dimnames(rec) <- dimnames(tsb) <- dimnames(x)
# One SAM table, arbitrary object name
sam2taf(x)
sam2taf(x, "SSB")
sam2taf(x, "SSB", year=FALSE)
# Many SAM tables, recognized names
sam2taf(rec)
data.frame(sam2taf(rec), sam2taf(tsb, year=FALSE))
## Not run:
# Accessing tables from SAM fit object
data.frame(sam2taf(rectable(fit)), sam2taf(tsbtable(fit), year=FALSE))
## End(Not run)
Run All TAF Scripts
Description
Run core TAF scripts in current directory.
Usage
source.all(...)
Arguments
... |
passed to |
Value
Logical vector, indicating which scripts ran without errors.
Note
TAF scripts that will be run if they exist are: utilities.R,
data.R, model.R, output.R, and report.R.
The model.R script may also be named method.R and is treated in
the same way.
See Also
source.taf runs a TAF script.
make.all runs all TAF scripts as needed.
clean cleans TAF directories.
TAF-package gives an overview of the package.
Examples
## Not run:
source.all()
## End(Not run)
Source Directory
Description
Read all *.R files from a directory containing R functions.
Usage
source.dir(dir, pattern = "\\.[r|R]$", all.files = FALSE,
recursive = FALSE, quiet = TRUE, ...)
Arguments
dir |
a directory containing R source files. |
pattern |
passed to |
all.files |
passed to |
recursive |
passed to |
quiet |
whether to suppress messages. |
... |
passed to |
Details
The dir argument can also be a vector of filenames, instead of a
directory name. This can be useful to specify certain files while avoiding
others.
Value
Names of sourced files.
Note
This function is convenient in TAF analyses when many R utility functions are stored in a directory, see example below.
See Also
source is the base function to read R code from a file.
TAF-package gives an overview of the package.
Examples
## Not run:
source.dir("boot/software/utilities")
## End(Not run)
Run TAF Script
Description
Run a TAF script and return to the original directory.
Usage
source.taf(script, rm = FALSE, clean = TRUE, detach = FALSE,
taf = NULL, quiet = FALSE)
Arguments
script |
script filename. |
rm |
whether to remove all objects from the global environment before and after the script is run. |
clean |
whether to |
detach |
whether to detach all non-base packages before running the script, to ensure that the script is not affected by packages that may have been attached outside the script. |
taf |
a convenience flag where |
quiet |
whether to suppress messages reporting progress. |
Details
The default value of rm = FALSE is to protect users from accidental
loss of work, but the TAF server always runs with rm = TRUE to make
sure that only files, not objects in memory, are carried over between
scripts.
Likewise, the TAF server runs with clean = TRUE to make sure that the
script starts with a clean directory. The target directory of a TAF script
has the same filename prefix as the script: data.R creates ‘data’
etc.
Value
TRUE or FALSE, indicating whether the script ran without
errors.
Note
Commands within a script (such as setwd) may change the working
directory, but source.taf guarantees that the working directory
reported by getwd() is the same before and after running a script.
See Also
source is the base function to run R scripts.
make.taf runs a TAF script if needed.
source.all runs all TAF scripts in a directory.
TAF-package gives an overview of the package.
Examples
## Not run:
write("print(pi)", "script.R")
source("script.R")
source.taf("script.R")
file.remove("script.R")
## End(Not run)
Summary Results in TAF Format
Description
Small summary results table to describe a TAF format data frame to store values by year.
Usage
summary.taf
Format
Data frame containing 16 columns:
Year | year |
Rec | recruitment, numbers at age 1 in this year (thousands) |
Rec_lo | lower 95% confidence limit |
Rec_hi | upper 95% confidence limit |
TSB | total stock biomass (tonnes) |
TSB_lo | lower 95% confidence limit |
TSB_hi | upper 95% confidence limit |
SSB | spawning stock biomass (tonnes) |
SSB_lo | lower 95% confidence limit |
SSB_hi | upper 95% confidence limit |
Removals | total removals, including catches due to unaccounted mortality |
Removals_lo | lower 95% confidence limit |
Removals_hi | upper 95% confidence limit |
Fbar | average fishing mortality (ages 2-4) |
Fbar_lo | lower 95% confidence limit |
Fbar_hi | upper 95% confidence limit |
Details
The data are an excerpt (first years) from the summary results table for North Sea cod from the ICES (2016) assessment.
Source
ICES (2016). Report of the working group on the assessment of demersal stocks in the North Sea and Skagerrak (WGNSSK). ICES CM 2016/ACOM:14, p. 673. doi:10.17895/ices.pub.5329.
See Also
div and rnd can modify a large number of columns.
TAF-package gives an overview of the package.
Examples
summary.taf
x <- div(summary.taf, "Rec|TSB|SSB|Removals", grep=TRUE)
x <- rnd(x, "Rec|TSB|SSB|Removals", grep=TRUE)
x <- rnd(x, "Fbar", 3, grep=TRUE)
Boot TAF Analysis
Description
Process metadata files ‘SOFTWARE.bib’ and ‘DATA.bib’ to set up software and data files required for the analysis.
Usage
taf.boot(software = TRUE, data = TRUE, clean = TRUE, force = FALSE,
taf = NULL, quiet = FALSE, ...)
Arguments
software |
whether to process |
data |
whether to process |
clean |
whether to |
force |
whether to remove existing |
taf |
a convenience flag where |
quiet |
whether to suppress messages reporting progress. |
... |
passed to |
Details
If clean = TRUE then:
-
clean.softwareandclean.libraryare run if ‘SOFTWARE.bib’ is processed. -
clean.datais run if ‘DATA.bib’ is processed.
The default behavior of taf.boot is to skip downloading of remote
files (GitHub resources, URLs, boot scripts) and also skip installing R
packages from GitHub if the files seem to be already in place. This is done
to speed up the boot procedure as much as possible. To override this and
guarantee that all data and software files are updated, pass force =
TRUE to download and install everything declared in SOFTWARE.bib and
DATA.bib.
Value
Logical vector indicating which metadata files were processed.
Note
This function should be called from the top directory of a TAF analysis. It looks for a directory called ‘boot’ and prepares data files and software according to metadata specifications.
The boot procedure consists of the following steps:
If a
boot/SOFTWARE.bibmetadata file exists, it is processed.If a
boot/DATA.bibmetadata file exists, it is processed.
After the boot procedure, software and data have been documented and are ready to be used in the subsequent analysis. Specifically, the procedure populates up to three new directories:
-
boot/datawith data files. -
boot/librarywith R packages compiled for the local platform. -
boot/softwarewith software files, such as R packages intar.gzsource code format.
From version 4.2 onwards, the term boot is preferred for what used to
be called bootstrap, mainly to avoid confusion with statistical
bootstrap. To taf.boot() is similar to booting a computer, readying
the components required for subsequent computations. Help pages now refer to
boot, but all TAF functions fully support existing analyses that have
a legacy bootstrap folder.
Model settings and configuration files can be set up within DATA.bib,
see TAF Wiki.
See Also
draft.data and draft.software can be used to
create initial draft versions of ‘DATA.bib’ and ‘SOFTWARE.bib’
metadata files.
taf.library loads a package from boot/library.
TAF-package gives an overview of the package.
Examples
## Not run:
taf.boot()
## End(Not run)
Construct Boot Path
Description
Construct a relative path to the boot folder, regardless of whether
the current working directory is the TAF root, the boot folder, or a
subfolder inside boot.
Usage
taf.boot.path(..., fsep = .Platform$file.sep)
Arguments
... |
names of folders or files to append to the result. |
fsep |
path separator to use instead of the default forward slash. |
Value
Relative path, or a vector of paths.
Note
This function is especially useful in boot scripts.
See Also
file.path is the underlying function used to construct the path.
taf.data.path constructs the path to boot data files.
TAF-package gives an overview of the package.
Examples
## Not run:
taf.boot.path()
taf.boot.path("software")
## End(Not run)
TAF Colors
Description
Predefined colors that can be useful in TAF plots.
Usage
taf.green
taf.orange
taf.blue
taf.dark
taf.light
See Also
TAF-package gives an overview of the package.
Examples
taf.green
opar <- par(mfrow=c(3,1))
barplot(5:1, main="Five",
col=c(taf.green, taf.orange, taf.blue, taf.dark, taf.light))
barplot(6:1, main="Six", col=c(taf.green, taf.orange, taf.blue,
taf.dark, taf.light, "white"))
barplot(7:1, main="Seven", col=c("black", taf.dark, taf.light,
taf.green, taf.orange, taf.blue, "white"))
par(opar)
Construct Boot Data Path
Description
Construct a relative path to data files in the boot data folder,
regardless of whether the current working directory is the TAF root, the
boot folder, or a subfolder inside boot.
Usage
taf.data.path(..., fsep = .Platform$file.sep)
Arguments
... |
filenames inside |
fsep |
path separator to use instead of the default forward slash. |
Value
Relative path, or a vector of paths.
Note
This function is especially useful in boot scripts.
See Also
file.path is the underlying function used to construct the path.
taf.boot.path constructs the path to the boot folder.
TAF-package gives an overview of the package.
Examples
taf.data.path()
taf.data.path("example.dat")
TAF Example
Description
Copy example analysis from TAF package.
Usage
taf.example(name, path = ".", force = FALSE)
Arguments
name |
of TAF example analysis. |
path |
where to create example directory. The default is the current working directory. |
force |
whether to overwrite existing directory. |
Details
Currently, the package comes with one example: "linreg".
Value
Full path to directory that was created.
Note
The example analysis is copied from the TAF package directory:
dir(system.file("examples", package="TAF"), full=TRUE)
See Also
taf.skeleton creates an empty TAF template.
linreg describes the linreg example.
TAF-package gives an overview of the package.
Examples
## Not run:
taf.example("linreg")
setwd("linreg")
taf.boot()
source.all()
## End(Not run)
TAF Install
Description
Install packages in ‘tar.gz’ format in local TAF library.
Usage
taf.install(targz = NULL, lib = taf.boot.path("library"), quiet = FALSE)
Arguments
targz |
a package filename, vector of filenames, or |
lib |
location of local TAF library. |
quiet |
whether to suppress messages. |
Details
If targz = NULL, all packages found in boot/software are
installed, as long as they have filenames of the form
package_sha.tar.gz containing a 7-character SHA reference code.
The default behavior of taf.install is to install packages in
alphabetical order. When the installation order matters because of
dependencies, the user can specify a vector of package filenames to install.
Value
No return value, called for side effects.
Note
The taf.boot procedure downloads and installs R packages, without
requiring the user to run taf.install. The main reason for a TAF user
to run taf.install directly is to initialize and run a TAF analysis
without running the boot procedure, e.g. to avoid updating the underlying
datasets and software.
After installing the package, this function writes the remote SHA reference
code into the package files DESCRIPTION and Meta/package.rds.
See Also
taf.boot calls download.github and
taf.install to download and install R packages.
taf.library loads a package from boot/library.
clean.library selectively removes packages from the local TAF
library.
install.packages is the underlying base function to install a
package.
TAF-package gives an overview of the package.
Examples
## Not run:
# Install one package
taf.install("boot/software/FLAssess_f1e5acb.tar.gz")
# Install all packages found in boot/software
taf.install()
## End(Not run)
Add TAF Library Path
Description
Add TAF library to the search path for R packages.
Usage
taf.libPaths(remove = FALSE)
Arguments
remove |
whether to remove TAF library from the search path, instead of adding it. |
Value
The resulting vector of file paths.
Warning
An unwanted side effect of having the TAF library as the first element in the
search path is that install.packages will then install packages inside
boot/library. This is not a serious side effect, since a subsequent
call to taf.boot or clean.library will remove packages from
the TAF library that are not declared in the ‘SOFTWARE.bib’ file.
Note
Specifically, this function sets "boot/library" as the first element
of .libPaths(). This is rarely beneficial in TAF scripts, but can be
useful when using the sessioninfo package, for example.
See Also
.libPaths is the underlying function to modify the search path
for R packages.
taf.library loads a package from boot/library.
TAF-package gives an overview of the package.
Examples
taf.libPaths()
taf.libPaths(remove=TRUE)
TAF Libraries
Description
Load and attach all packages from local TAF library.
Usage
taf.libraries(messages = FALSE, warnings = FALSE)
Arguments
messages |
whether to show messages when package loads. |
warnings |
whether to show warnings when package loads. |
Value
TRUE (invisibly) if all packages loaded.
Note
Packages in the TAF library are loaded in the order in which they are
listed in SOFTWARE.bib. Internal dependencies can in this way be
respected.
See Also
taf.library is the TAF function called for each found package.
TAF-package gives an overview of the package.
Examples
## Not run:
# Load all packages in TAF library
taf.libraries()
## End(Not run)
TAF Library
Description
Load and attach package from local TAF library.
Usage
taf.library(package, messages = FALSE, warnings = FALSE)
Arguments
package |
name of a package found in |
messages |
whether to show messages when package loads. |
warnings |
whether to show warnings when package loads. |
Value
The names of packages currently installed in the TAF library.
Note
The purpose of the TAF library is to retain R packages that are not commonly used (and not on CRAN), to support long-term reproducibility of TAF analyses.
If a package has dependencies that are also in the TAF library, they will be
loaded in preference of any version that may be installed in the system or
user library. To force the use of a dependency from outside of the TAF
library call library(package) prior to the call to taf.library.
See Also
library is the underlying base function to load and attach a
package.
taf.boot is the procedure to install packages into a local TAF
library, via the SOFTWARE.bib metadata file.
detach.packages detaches all packages.
TAF-package gives an overview of the package.
Examples
## Not run:
# Show packages in TAF library
taf.library()
# Load packages
taf.library(this)
taf.library(that)
## End(Not run)
PNG Device
Description
Open PNG graphics device to export a plot into the TAF report folder.
Usage
taf.png(filename, width = 1600, height = 1200, res = 200, ...)
Arguments
filename |
plot filename. |
width |
image width. |
height |
image height. |
res |
resolution determining the text size, line width, plot symbol size, etc. |
... |
passed to |
Details
The filename can be passed without the preceding
"report/", and without the ".png" filename extension.
Specifically, the function prepends "report/" to the filename if (1)
the filename does not contain a "/" separator, (2) the working
directory is not report, and (3) the directory report exists.
The function also appends ".png" to the filename if it does not
already have that filename extension.
This automatic filename manipulation can be bypassed by using the png
function directly.
Value
No return value, called for side effects.
Note
A simple convenience function to shorten
png("report/plot.png", width=1600, height=1200, res=200)
to
taf.png("plot")
The res argument affects the text size, along with all other plot
elements. To change the text size of specific lattice plot elements, the
zoom function can be helpful.
For consistent image width and text size, it can be useful to keep the
default width = 1600 but vary the height to adjust the desired
aspect ratio for each plot.
See Also
png is the underlying function used to open a PNG graphics
device.
zoom changes text size in a lattice plot.
TAF-package gives an overview of the package.
Examples
## Not run:
taf.png("myplot")
plot(1)
dev.off()
library(lattice)
taf.png("mytrellis")
xyplot(1~1)
dev.off()
library(ggplot2)
taf.png("myggplot")
qplot(1, 1)
dev.off()
## End(Not run)
TAF Session
Description
Show session information about loaded packages, clearly indicating which packages were loaded from the local TAF library.
Usage
taf.session(sort = FALSE, imports = TRUE, details = FALSE)
Arguments
sort |
whether to sort packages by name. |
imports |
whether to include imported packages. |
details |
whether to report more detailed session information. |
Value
List containing session information about loaded packages.
See Also
sessionInfo and the sessioninfo package provide similar
information, but do not indicate clearly packages that were loaded from the
local TAF library.
TAF-package gives an overview of the package.
Examples
taf.session()
taf.session(sort=TRUE)
taf.session(imports=FALSE)
taf.session(details=TRUE)
TAF Skeleton
Description
Create initial directories and R scripts for a new TAF analysis.
Usage
taf.skeleton(path = ".", force = FALSE, pkgs = taf.pkg(),
model.script = "model.R", gitignore = TRUE)
Arguments
path |
where to create initial directories and R scripts. The default is the current working directory. |
force |
whether to overwrite existing scripts. |
pkgs |
packages to load at the start of each script. The default is
either |
model.script |
model script filename, either |
gitignore |
whether to write TAF entries to a ‘.gitignore’ file. |
Details
When gitignore = TRUE, the following entries will be written to a
.gitignore file, appending if the file exists already:
/boot/data /boot/library /boot/software /data /model /output /report *.Rproj .RData .Rhistory .Rproj.user .Ruserdata
Value
Full path to analysis directory.
Note
After running taf.skeleton() to create a new TAF workflow, the author
can populate the boot/initial/data folder with initial data files and
run draft.data(file=TRUE) to produce a DATA.bib file.
The next step is then to run taf.boot() to populate the
boot/data folder and start editing the data.R script, reading
files from the boot/data folder.
See Also
taf.example copies an example analysis from the TAF package.
package.skeleton creates an empty template for a new R package.
TAF-package gives an overview of the package.
Examples
## Not run:
taf.skeleton()
## End(Not run)
List Sources
Description
List metadata entries from DATA.bib, SOFTWARE.bib, or both.
Usage
taf.sources(type)
Arguments
type |
one of |
Value
List of metadata entries.
Note
The functionality is similar to read.bib, with the addition of a
type field, indicating whether an entry is data
software.
This function is used internally by the taf.boot procedure and is
also useful when organizing a larger TAF project.
See Also
taf.boot reads and processes metadata entries.
read.bib is the underlying function to read metadata entries.
process.entry processes a single metadata entry, in the list
format returned by taf.sources.
Examples
## Not run:
taf.sources("data")
taf.sources("software")
taf.sources("both")
## End(Not run)
Unzip File
Description
Extract files from a zip archive, retaining executable file permissions.
Usage
taf.unzip(zipfile, files = NULL, exdir = ".", unzip = NULL, ...)
Arguments
zipfile |
zip archive filename. |
files |
files to extract, default is all files. |
exdir |
directory to extract to, will be created if necessary. |
unzip |
extraction method to use, see details below. |
... |
passed to |
Details
The default method unzip = NULL uses the external unzip
program in Unix-compatible operating systems, but an internal method in
Windows. For additional information, see the unzip help page.
Value
No return value, called for side effects.
Note
One shortcoming of the base unzip function is that the default
"internal" method resets file permissions, so Linux and macOS
executables will return a 'Permission denied' error when run.
This function is identical to the base unzip function, except the
default value unzip = NULL chooses an appropriate extraction method in
all operating systems, making it useful when writing platform-independent
scripts.
See Also
unzip is the base function to unzip files.
TAF-package gives an overview of the package.
Examples
## Not run:
exefile <- if(os.unix()) "run" else "run.exe"
taf.unzip("boot/software/archive.zip", files=exefile, exdir="model")
## End(Not run)
Convert TAF Table to HTML
Description
Convert a data frame to HTML code and optionally write to a file.
Usage
taf2html(x, file = "", align = "", header = align,
digits = getOption("digits"), center = "style=\"text-align:center\"",
left = "style=\"text-align:left\"",
right = "style=\"text-align:right\"", append = FALSE)
Arguments
x |
a data frame. |
file |
a filename, or special value |
align |
a string (or a vector of strings) specifying alignment of data cells. |
header |
a string (or a vector strings) specifying alignment of header cells. |
digits |
significant digits for numeric columns. |
center |
HTML attribute to indicate center alignment. |
left |
HTML attribute to indicate left alignment. |
right |
HTML attribute to indicate right alignment. |
append |
whether to append to an existing file. |
Details
The align argument can be a vector of strings to specify
column-specific alignment, for example c("l","r","l","l"). Only the
first letter (case-insensitive) is used, so "left" is equivalent to
"L". An empty string (the default), or any string that does not begin
with C, L, or R indicates no specific alignment.
The header argument can be used to specify an alignment for the column
names that is different from the data values. The default is to use the same
alignment as the data values.
The center, left, and right arguments can be used to
specify the exact HTML attribute to render alignment, for users who are
familiar with cascading style sheets (CSS). For example, the long-winded
style="text-align:center" could be shortened to class="L" if a
corresponding class has been defined in CSS.
The default value file = "" prints the HTML code in the console,
instead of writing it to a file. The output can then be pasted into a file to
edit further, without accidentally overwriting an existing file.
Value
Character vector of class Bibtex.
Note
Although the output is HTML code, the Bibtex class is used for
convenient display in the console.
The resulting HTML conforms to the HTML5 standard and aims for compact output, omitting optional closing tags and rendering each row of data as one row of HTML code.
See Also
write.taf writes a data frame to a CSV file.
TAF-package gives an overview of the package.
Examples
taf2html(catage.taf)
taf2html(catage.taf, align=c("L","R","R","R","R"))
## Not run:
taf2html(catage.taf, "catage.html")
taf2html(catage.taf, "catage.html", align=c("L","R","R","R","R"),
append=TRUE)
## End(Not run)
Convert TAF Table to Long Format
Description
Convert a table from TAF format to long format.
Usage
taf2long(x, names = c("Year", "Age", "Value"))
Arguments
x |
a data frame in TAF format. |
names |
a vector of three column names for the resulting data frame. |
Value
A data frame with three columns.
Note
TAF stores tables as data frames, usually with a year column as seen in stock assessment reports. The long format is more convenient for analysis and producing plots.
See Also
catage.taf and catage.long describe the TAF and
long formats.
long2taf converts a long table to TAF format.
wide2long converts a wide table to long format.
TAF-package gives an overview of the package.
Examples
taf2long(catage.taf, names=c("Year","Age","Catch"))
Convert TAF Table to Crosstab Format
Description
Convert a table from TAF format to crosstab format.
Usage
taf2xtab(x)
Arguments
x |
a data frame in TAF format. |
Value
A data frame with years as row names.
Note
TAF stores tables as data frames, usually with a year column as seen in stock assessment reports. The crosstab format can be more convenient for analysis and producing plots.
See Also
catage.taf and catage.xtab describe the TAF and
crosstab formats.
tt converts a TAF table to transposed crosstab format.
xtab2taf converts a crosstab table to TAF format.
TAF-package gives an overview of the package.
Examples
taf2xtab(catage.taf)
TAF Transpose
Description
Convert a table from TAF format to transposed crosstab format.
Usage
tt(x, column = FALSE)
Arguments
x |
a data frame in TAF format. |
column |
a logical indicating whether the group names should be stored
in a column called ‘Age’ instead of in row names. Alternatively,
|
Value
A data frame with years as column names.
Note
Transposing can be useful when comparing TAF tables to stock assessment reports.
See Also
t transposes a matrix.
catage.taf describes the TAF format.
taf2xtab converts a TAF table to crosstab format, without
transposing.
TAF-package gives an overview of the package.
Examples
taf2xtab(catage.taf)
tt(catage.taf)
tt(catage.taf, TRUE)
tt(catage.taf, "Custom")
Convert Wide Table to Long Format
Description
Convert a table from wide format to long format.
Usage
wide2long(x, names = c("Age", "Value"))
Arguments
x |
a data frame in wide format. |
names |
a vector of two names for the last two columns of the resulting data frame. |
Value
A data frame.
Note
TAF stores tables as data frames, usually with a year column as seen in stock
assessment reports. Although year is the most common dimension, tables may
also include area, sex, season, or other additional dimensions. The
catage.wide table provides an example of a wide table that includes
area and year as dimensions. The long format is more convenient for analysis
and producing plots.
See Also
catage.taf and catage.wide describe the TAF and
wide formats.
taf2long converts a TAF table to long format.
TAF-package gives an overview of the package.
Examples
wide2long(catage.wide, names=c("Age","Catch"))
Write TAF Table
Description
Write a data frame to a CSV file.
Usage
write.taf(x, file = NULL, dir = NULL, quote = FALSE, row.names = FALSE,
fileEncoding = "UTF-8", underscore = TRUE, ...)
Arguments
x |
a data frame. |
file |
a filename. |
dir |
an optional directory name. |
quote |
whether to quote strings. |
row.names |
whether to include row names. |
fileEncoding |
character encoding for output file. |
underscore |
whether automatically generated filenames (when
|
... |
passed to |
Details
Alternatively, x can be a list of data frames or a string vector of
object names, to write many tables in one call. The resulting files are named
automatically, similar to file = NULL.
The default value file = NULL uses the name of x as a filename,
so a data frame called survey.uk will be written to a file called
‘survey_uk.csv’ (when underscore = TRUE) or ‘survey.uk.csv’
(when underscore = FALSE).
The special value file = "" prints the data frame in the console,
similar to write.csv.
Value
No return value, called for side effects.
Note
This function gives a warning when column names are missing or duplicated,
unless the target directory name is report. It also gives a warning if
the data frame has zero rows.
See Also
write.csv is the underlying function used to write a table to a
file.
read.taf reads from a CSV file into a data frame.
taf2html converts a data frame to HTML.
TAF-package gives an overview of the package.
Examples
## Not run:
write.taf(catage.taf, "catage.csv")
catage <- read.taf("catage.csv")
write.taf(catage)
file.remove("catage.csv")
## End(Not run)
Convert Crosstab Table to Long Format
Description
Convert a table from crosstab format to long format.
Usage
xtab2long(x, names = c("Year", "Age", "Value"))
Arguments
x |
a data frame in crosstab format. |
names |
a vector of three column names for the resulting data frame. |
Value
A data frame with three columns.
See Also
catage.xtab and catage.long describe the crosstab
and long formats.
xtab2taf and taf2long are the underlying
functions that perform the conversion.
TAF-package gives an overview of the package.
Examples
xtab2long(catage.xtab, names=c("Year","Age","Catch"))
Convert Crosstab Table to TAF Format
Description
Convert a table from crosstab format to TAF format.
Usage
xtab2taf(x, colname = "Year")
Arguments
x |
a data frame in crosstab format. |
colname |
name for first column. |
Value
A data frame in TAF format.
Note
TAF stores tables as data frames, usually with a year column as seen in stock assessment reports. The crosstab format can be more convenient for analysis and producing plots.
See Also
catage.xtab and catage.taf describe the crosstab
and TAF formats.
taf2xtab converts a TAF table to crosstab format.
TAF-package gives an overview of the package.
Examples
xtab2taf(catage.xtab)
Zoom
Description
Change text size in a lattice plot.
Usage
zoom(x, ...)
## S3 method for class 'trellis'
zoom(x, size = 1, main = 1.2 * size, lab = size,
axis = size, strip = size, sub = 0.9 * size, legend = 0.9 * size,
splom = 0.9 * size, ...)
Arguments
x |
a lattice plot of class |
... |
further arguments, currently ignored. |
size |
text size multiplier. |
main |
size of main title (default is |
lab |
size of axis labels (default is |
axis |
size of tick labels (default is |
strip |
size of strip labels (default is |
sub |
size of subtitle (default is |
legend |
size of legend labels (default is |
splom |
size of scatterplot matrix diagonal labels (default is
|
Details
Pass NULL for any argument to avoid changing the size of that text
component.
The legend component of a lattice plot can be somewhat fickle, as the
object structure varies between plots. One solution is to pass
legend = NULL and tweak the legend before or after calling the
zoom function.
Value
The same lattice object, but with altered text size.
Note
The default values result in lattice plots that have similar text size as
base plots, when using taf.png.
This function ends with a print call, to
make it easy to export the lattice plot to a file, without the need of an
explicit print.
See Also
Lattice plots are created using
xyplot or related functions.
taf.png opens a PNG graphics device.
TAF-package gives an overview of the package.
Examples
library(lattice)
xyplot(1~1)
zoom(xyplot(1~1))
zoom(xyplot(1~1), size=1.2)
zoom(xyplot(1~1), axis=0.8)
zoom(xyplot(1~1), axis=NULL)
## Not run:
taf.png("myplot")
plot(1)
dev.off()
taf.png("mytrellis")
xyplot(1~1)
dev.off()
taf.png("mytrellis_zoom")
zoom(xyplot(1~1))
dev.off()
## End(Not run)