






This package provides functions for fitting Horseshoe Trees, Causal Horseshoe Forests, and their more general counterparts: Shrinkage Trees and Causal Shrinkage Forests.
These models allow for global-local shrinkage priors on tree step heights, enabling adaptive modeling in high-dimensional settings.
The functions can be used for:
Supported outcome types: continuous, binary, and right-censored survival times.
The mathematical background and theoretical foundation for these models is described in the preprint Horseshoe Forests for High-Dimensional Causal Survival Analysis by T. Jacobs, W.N. van Wieringen, and S.L. van der Pas (arXiv:2507.22004).
The released version of ShrinkageTrees can be installed from CRAN:
install.packages("ShrinkageTrees")You can install the development version from GitHub:
# Install devtools if not already installed
install.packages("devtools")
devtools::install_github("tijn-jacobs/ShrinkageTrees")library(ShrinkageTrees)
set.seed(42)
n <- 100
p <- 1000
# Generate covariates
X <- matrix(runif(n * p), ncol = p)
X_treat <- X_control <- X
treatment <- rbinom(n, 1, X[, 1])
tau <- 1 + X[, 2]/2 - X[, 3]/3 + X[, 4]/4
# Generate survival times (on log-scale)
true_time <- X[, 1] + treatment * tau + rnorm(n)
censor_time <- log(rexp(n, rate = 0.05))
follow_up <- pmin(true_time, censor_time)
status <- as.integer(true_time <= censor_time)
# Fit a standard Causal Horseshoe Forest (without propensity score adjustment)
fit_horseshoe <- CausalHorseForest(
y = follow_up,
status = status,
X_train_control = X_control,
X_train_treat = X_treat,
treatment_indicator_train = treatment,
outcome_type = "right-censored",
timescale = "log",
number_of_trees = 200,
k = 0.1,
N_post = 5000,
N_burn = 5000,
store_posterior_sample = TRUE
)
# Posterior mean CATEs
CATE_horseshoe <- colMeans(fit_horseshoe$train_predictions_sample_treat)
# Posteriors of the ATE
post_ATE_horseshoe <- rowMeans(fit_horseshoe$train_predictions_sample_treat)
# Posterior mean ATE
ATE_horseshoe <- mean(post_ATE_horseshoe)
# Plot the posterior of the ATE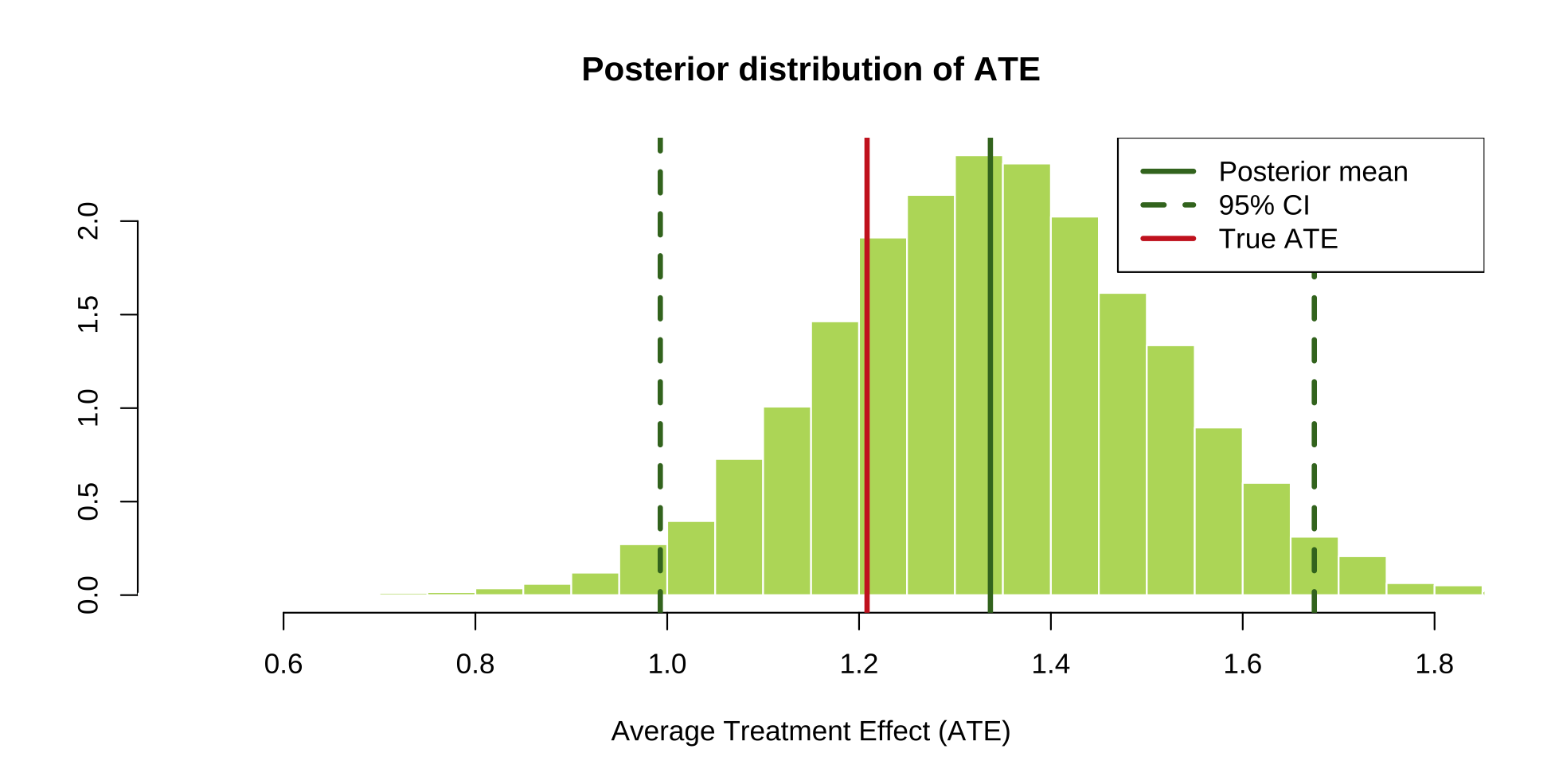
The package includes a demo analysis based on the TCGA PAAD (pancreatic cancer) dataset to showcase how ShrinkageTrees can be used in practice. This demo replicates the main case study from the preprint “Horseshoe Forests for High-Dimensional Causal Survival Analysis” (arXiv:2507.22004).
The demo: - Estimates propensity scores for treatment
assignment
- Fits a Causal Horseshoe Forest to the survival times with
right-censoring
- Computes the posterior mean ATE and individual CATEs with 95% credible
intervals
- Produces diagnostic plots (propensity score overlap, posterior ATE,
CATE estimates, sigma trace)
You can run it directly from R after installing the package:
demo("pdac_analysis", package = "ShrinkageTrees")?ShrinkageTrees, ?HorseTrees,
?CausalHorseForest, and ?CausalShrinkageForest
for detailed help.Contributions are welcome! Feel free to open an issue or submit a pull request. The software is designed to be flexible and modular, allowing for a wide variety of global-local shrinkage priors to be easily implemented and extended in future versions.
This package is licensed under the MIT License.
This project has received funding from the European Research Council (ERC) under the European Union’s Horizon Europe program under Grant agreement No. 101074082. Views and opinions expressed are however those of the author(s) only and do not necessarily reflect those of the European Union or the European Research Council Executive Agency. Neither the European Union nor the granting authority can be held responsible for them